Metabolomics for Biomarker Discovery
Recent advances in non-targeted metabolite profiling (i.e., metabolomics) offers a revolutionary approach for phenotyping individuals at a molecular level that is needed for new breakthroughs in precision medicine. Similar to the genome, the “metabolome” consists of all small molecules (< 1.5 kDa) that define the functional activity of a cell or organism. Metabolites not only refer to endogenous by-products of cellular metabolism, but also xenobiotics and bioactive compounds derived from diet, environment and gut microflora. Since metabolites are “real-world”end-products ofgeneexpression and environmental exposures,metabolomics offers a promising approach for biomarker discovery. Our group is interested in developing an accelerated data workflow in metabolomics that take advantage of multiplexed separations based on CE-MS for biomarker discovery with high data fidelity. In conjunction with multivariate statistical analysis for the identification and quantification of unknown metabolites in complex biological samples. This work has promising implications for contributing new advances in personalized medicine, drug development and the early detection of inherited and chronic diseases. For instance, his recent contributions have identified novel sweat biomarkers of cystic fibrosis for confirmatory testing of presumptive screen positive infants in support of universal newborn screening programs.
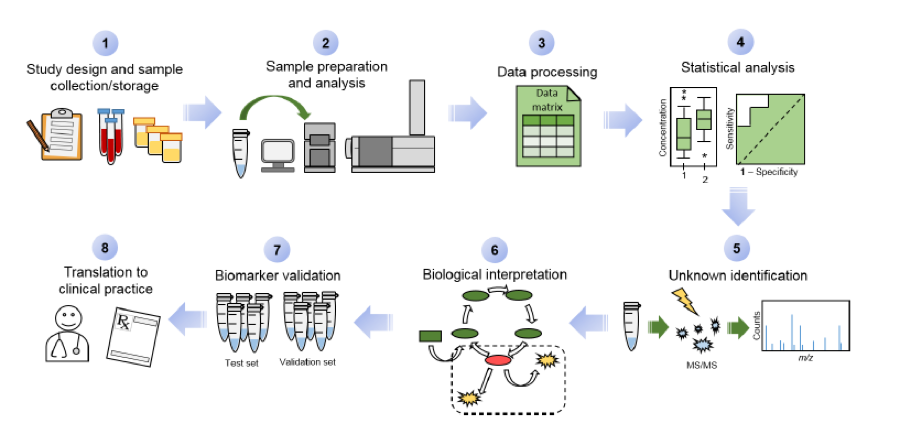
Fig. 1. Overview of the data workflow in metabolomics for the discovery and validation of diagnostic, prognostic or predictive biomarkers relevant to human diseases in clinical medicine.
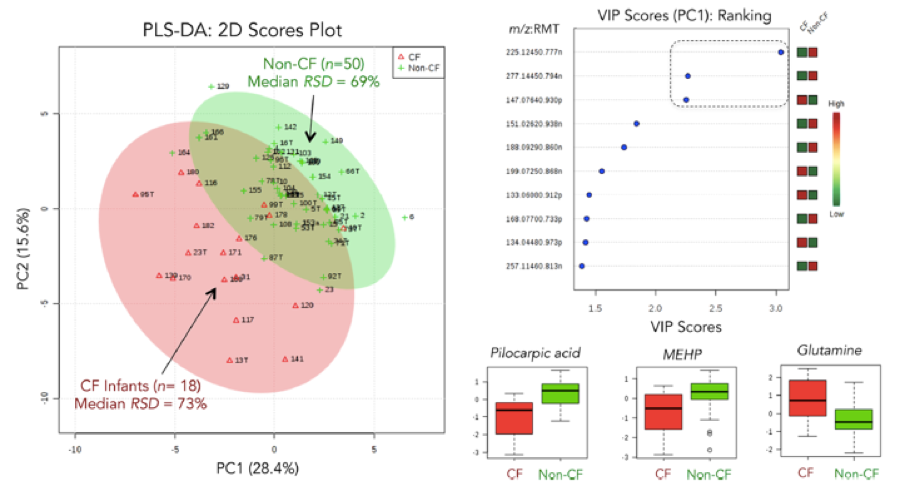
Fig. 2. Sweat metabolomics for the identification of novel biomarkers that differentiate asymptomatic cystic fibrosis (CF) infants from unaffected carriers beyond sweat chloride, including two unexpected exogenous metabolites, namely pilocarpic acid and monoethylhexylphthalate (MEHP) that likely reflect a higher risk for paraoxanase deficiency in CF.
Recommended References:
- A. DiBattista,* N. McIntosh, M. Lamoureux, O.Y. Al-Dirbashi, P. Chakraborty and P. Britz-McKibbin “Metabolic Signatures of Cystic Fibrosis Identified in Dried Blood Spots for Newborn Screening Without Carrier Identification” J. Proteome Res. 2019, 18, 841–854.
- A. Nori de Macedo,* S. Mathiaparanam,* L. Brick, K. Keenan, T. Gonska, L. Pedder, S. Hill, and P. Britz-McKibbin “Characterization of the Sweat Metabolome in Screen-Positive Cystic Fibrosis Infants: Revealing Mechanisms Beyond Impaired Chloride Transport” ACS Central Science 2017, 3: 904-914.
- N.L. Kuehnbaum,* A. Kormendi*, A. DiBattista,* K.P. Lam,* J.B. Gillen,* M.J. Gibala and P. Britz-McKibbin “Multiplexed Separations for Biomarker Discovery in Metabolomics: Elucidating Adaptive Responses to Exercise Training” Electrophoresis 2015, 36:2226-2236.
- F. Badoud,* K. P. Lam,* M. Perreault, M.A. Zulyniak, P. Britz-McKibbin and D. Mutch, “Metabolomics Reveals Metabolically Healthy and Unhealthy Obese Individuals Differ in their Response to a Caloric Challenge” PLoS One 2015, 10:e0134613.
